Review Article
Impact of Orthodontic Force on Genes During Leveling and Alignment: A Systematic Review
1168
Views & Citations168
Likes & Shares
Background: Orthodontic tooth movement is possible as cementum is more resistant to resorption. Bone remodeling is a continuous process throughout. Several theories have been proposed to explain the biology of tooth movement. Gene and gene therapy is gaining more importance in explaining the orthodontic tooth movement.
Objective: The objective of this systematic review is to decisively appraise the possible effect of orthodontic force on genes during leveling and alignment.
Materials and methods: A literature assessment was performed using electronic databases. The search was made using PubMed, www.ncbi.nim.nih.gov, Scupose, Hinary, Ebscodatabase search engines. The assessment of review of literature enclosed from January 1970 to December 2019 and used the MeSH term “role of genes in tooth leveling and alignment, orthodontic forces, and gene reaction during leveling and alignment.
Selection criteria: PRISMA analytic selection criteria have been applied to select the studies that reported on orthodontic force and its effects on genes during leveling and alignment. Only randomized control trials, descriptive, diagnostic, cross-sectional and longitudinal types of studies were selected.
Data collection and analysis: The data extracted from the selected articles were based on year of publication, study design, age of subjects, specific type gene involved, and author’s conclusions. A quality assessment of the procedural accuracy of each article was executed as per the method described by Jadad et al for RCT, metal analysis, CCT, cross-sectional studies, longitudinal and retrospective studies.
Results: Literature search identified 117 records from electronic databases and the partial grey literature (Google scholar) search. Finally, 8 articles fulfilled the eligibility criteria were included in the review.
Limitations: All the reviewed studies were of the high risk of error due to smaller sample size and they are cross-sectional in studies.
Conclusions: This systematic review concludes answering the question that on the application of a biomechanical stimulus, initiates the release of IL1, then activates the nociceptive pain pathway followed by up and down regulatory pathways in presence of TNF α, M-CSF, and TGFβ.
Keywords: Orthodontics, Genes, Biomechanical, Tooth movements, Bone
Abbreviations: OTM: Orthodontic Tooth Movement; CGRP: Calcitonin Gene Related Peptide; VEGF: Vascular Endothelial Growth Factor; IL: Inter Leukane; PGE: Prostaglandines E2; TNF α: Tumour Necrosis Factor alpha; M-CSF: Macrophage-Colony Stimulating Factor; PTHrp: Parathyroid Hormone-Related Protein; IHH: Indian Hedgehog; MMPs: Matrix metalloproteinases; TIMPs: Tissue inhibitor of Matrix metalloproteinases; OPG: Osteoprotegerin; G-CSF: Granulocytes Colony-Stimulating Factor
INTRODUCTION
Development begins with the legacy of genes variation. The term genes can be defined as the basic physical and functional unit of heredity, composed of DNA; act as instructions to make molecules called proteins. Genetic factors control various biological processes including orthodontic tooth movement (OTM). Comprehensive knowledge of genetic factors and their associated problems with OTM, treatment outcomes can be enhanced. Hence, if orthodontic tooth movement is a play, in the biological stage of PDL, genes are the directors, guides the rest of the cells of PDL to places teeth in proper leveling and alignment [1-5].
The orthodontic force generates and propagates the signaling cascade molecule. In response to applied mechanical load bone remodeling initiates in adjacent tissue. This forms the central theme of the orthodontic tooth movement. The aptitude of adaptive reaction to applied orthodontic force is coded in the DNA of periodontal ligament (PDL) and alveolar bone cells. It is the fundamental assets of any cell to react and determine the molecular genetic response making tooth movement possible [5-9].
In general, orthodontic tooth movement can vary depending on the nature of the applied force and reaction of PDL. The orthodontic force alters the vascularity and blood flow of PDL resulting in the release of significant molecules like neurotransmitters, cytokines, growth factors, colony-stimulating factors, and arachidonic acid metabolites. These molecules can induce various cellular responses in and around teeth, creating a suitable microenvironment for tissue remodeling [8-11].
The PDL is a specialized tissue that plays an essential role in signal transduction pathways concerning repair and remodeling of the PL, cementum, and alveolar bone. Homeobox protein MSX 2 present in periodontal ligament as a molecular defense mechanism. During orthodontic tooth movement, it prevents tooth ankylosis. The PDL cellular matrix such as Fibroblasts, osteoblasts, osteocytes, osteoclasts, odontoblasts, cementoblasts, and chondrocytes are involved in the remodeling process [1-18].
To date, current literature has assorted information on genetic-level cellular responses to orthodontic force. The bone cell is linked to various genes, on the application of orthodontic force bone cell releases various enzymes like glutamate/ aspartate transporter, inducible nitric oxide synthase, and prostaglandin G/H synthetase. For example, on the application of orthodontic force, the bone Releases osteopontin mRNA within 12 h and retains for 48 h. Various researchers have suggested that 26 genes are implicated in osteoclast release. These are tyrosine kinase gene, M-CSF, C-fos, Pu.1, and NF-_B (osteoclast formation), and C-tyrosine kinase and microphthalmia transcription factor in bone remodeling process [18-23].
This systematic review aimed to find an answer to the question that ‘how orthodontic force does activate genes during orthodontic tooth movement’.
MATERIALS & METHODS
The systematic review put up with by The Preferred Reporting Item for Systematic Review and Meta-Analysis (PRISMA) checklist reporting tool.
Protocol registration
This study protocol was not registered.
Eligibility criteria (Table 1)
The studies included were on orthodontic force and its effects on genes during leveling and alignment. Those are randomized control trials, Descriptive; diagnostic, cross-sectional and longitudinal types of studies.
No age or gender restrictions were applied.
Exclusion criteria (Table 1): Included studies that did not report role of genes during orthodontic tooth movement outcomes.
CASE REPORTS
Information sources
A comprehensive systematic search of the following electronic database was executed up to 2020: PubMed, Medline (Ovid), Cochrane Library, Scopus, EMBASE (OvidSP) and Google Scholar.
Search strategy
The search approach was planned with the support of a health sciences librarian. The search was conducted using keywords orthodontic force and its effects on genes during leveling and alignment, a combination of keywords ‘role of genes in orthodontic tooth movements with truncation, and medical subject headings. (MESH). A grey literature exploration was performed with Google Scholar and most relevant articles were selected. The investigating tactics for Medline (OvidSP), Embase (OvidSP), PubMed, Scopus, Cochrane, was modified to assist in pointing out other databases. All references were managed by using reference manager software and duplicates were removed.
Study selection
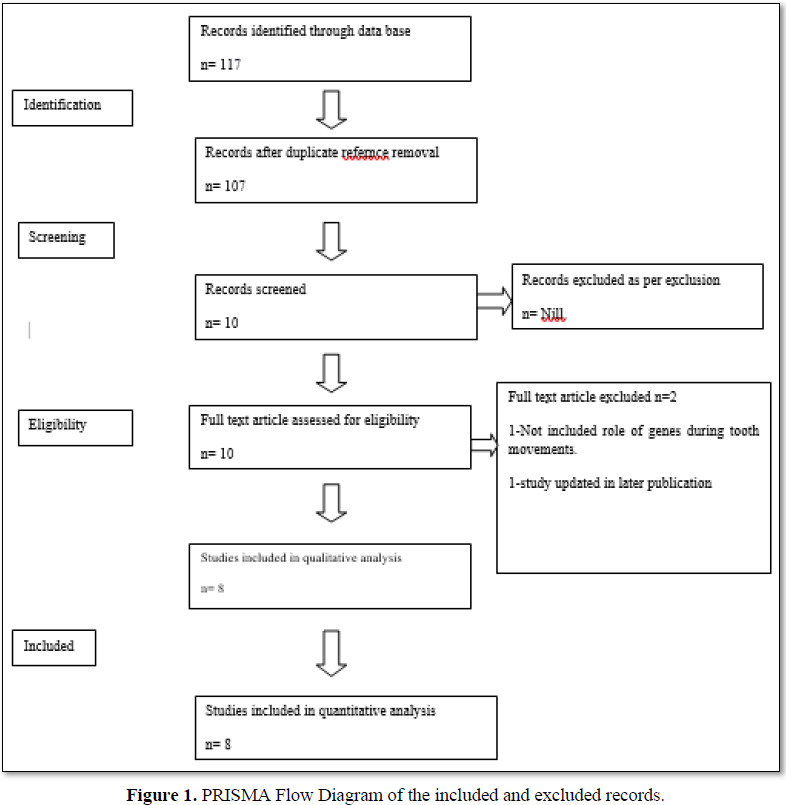
Study assortment was accomplished in two phases. In the first phase, 2 reviewers (UR and RC) separately screened the titles and abstracts acknowledged by all electronic databases related to the Effects of orthodontic force on genes during leveling and alignment. In the second phase, full-text articles were evaluated by reviewers to confirm their final selection. Cohen’s kappa coefficient was calculated to quantify the reviewer conformity in the study selection process (Figure 1).
Data collection process and data items
The same two reviewers (UR and RC) autonomously performed data collection and scrutinized all information to validate the completeness of the retrieved data. The data extraction integrated the following items: General information (name of the authors, the year of publication and study setting); methods (study design, comparison groups and periodontal assessment method); sample size; and outcomes of each study (Table 2).
RESULTS
Study selection
The literature search identified 117 records from electronic databases and the partial grey literature (Google scholar) search. Duplicate references were expelled and a total of 107 articles were meticulously analyzed. The reviewer agreement (Kappa) was considered by assessing the chosen titles and abstracts. The typical values for all databases were 0.96, which recommended a high level of agreement between reviewers. After analyzing the titles/abstracts and according to the inclusion criteria, 51 prospectively related studies were examined in depth. Finally, eight articles were included for qualitative analysis (Figure 1).
Study characteristics
Out of the eight included studies, three were RCTs. Between these eight (3) studies, three articles Effects of orthodontic force on genes, two (2) articles were on orthodontic tooth movement and ECM remodeling, one (01) study compared the role of cytokines, growth factors and transcription factors facilitated orthodontic tooth movement, and one study was on RANKL/OPG pathway, (02) two studies were on the effects of orthodontic force on various genetic receptors.
Risk of bias assessment
The outcomes of risk of bias consideration within studies are presented using the A quality assessment of the procedural accuracy of each article was executed as per the method described for RCT, metal analysis, CCT, cross-sectional studies, longitudinal and retrospective studies. Only three (3) of the eight preferred RCTs effectively deal with sequence generation (randomization). The additional five studies did not specify the sampling procedure. Statistical analysis was not performed in five of the studies.
Synthesis of results
Seven out of the eight studies scrutinized Orthodontic tooth movement and ECM remodeling with few genes’ intervention, three of them assessed the pivotal role of various genes in different phases of orthodontic teeth movement, and another two explored various receptors, two studies were the role of cytokines, growth factors and transcription factors and one study was demonstrated Pressure: Tension Related Effects and RANK RANKL/OPG pathway.
Orthodontic tooth movement and ECM remodeling
The orthodontic force induces biomechanical strain in PDL and dental pulp. This process displaces the ECM fluid in alveolar bone canaliculi and activates inflammation-mediated nociceptive pain [1-23]. The activation of the nociceptive pain pathway secretes neuropeptides such as Substance P, calcitonin gene-related peptide (CGRP) 23-26. The release of CGRP activates the release of vascular endothelial growth factor (VEGF) plays a pivotal role in new bone development. The newly formed bone reorganized through activation of the kinase- pathway via synthesis of extracellular signal-regulated kinase and intracellular proteins [1-26].
CYTOKINES IN ORTHODONTIC TOOTH MOVEMENT
Cytokines are the immunomodulating mediator mediates in autocrine, paracrine, and endocrine signaling to target cells in cell-cell fashion communication. This process modulates cell growth, proliferation, cell migration, differentiation, gene expression and cell-specific functions.
To date, various types of pro-inflammatory cytokines (IL-1 β, Il-6, IL-8, Il-12, IL-13 TNF-alpha), anti-inflammatory cytokine IL-1 is seen in both compressed and tension zone. The applied orthodontic force indirectly induces the secretion IL1. The released IL1 activates the PGE2 pathway and bone resorption in pressure occurs. Thus, the secretion of pro-inflammatory cytokines especially IL1 stimulates osteoclastic activity in the compressed side.
Recent studies have shown that IL1 is a potent by-product of the Tumour Necrosis Factor-alpha (TNF α) molecule. The mechanism of bone resorption in the compressed side is due to a diverse range of signaling events within cells in response to orthodontic force. For example, the activated TNFα in the presence of macrophage colony-stimulating factor (M-CSF) activates osteoclast progenitors to osteoclasts in the compressed side [1-23,27-31].
PRESSURE: TENSION RELATED EFFECTS
In response to biomechanical stimuli, two zones are created in the periodontal ligament. First, the tension zone occurs due to the activation of the kinase pathway. Second, inflammation-mediated nociceptive pain pathway activates IL1 in the presence of macrophage colony-stimulating factor (M-CSF) activates osteoclast progenitors to osteoclasts in compressed side resorption occurs.
The process of orthodontic tooth movement is regulated by parathyroid hormone-related protein (PTHrp) and Indian Hedgehog (IHH) directly in regulating the Type II collagen genes. In growing children growth modification is achieved PTHrP pathway regulating through SOX-9 and Indian Hedgehog (IHH) genes.
To date, in the growth modulating process Indian Hedgehog (IHH) brings the skeletal changes. Tooth movement is regulated by Matrix metalloproteinases (MMPs) with their Tissue inhibitor of Matrix metalloproteinases (TIMPs) [1-23,27-33].
Rank Rankl/Opg Pathway [11-18]
Osteoprotegerin (OPG) is fundamental glycoproteins consist of 401 amino acid residues arranged as7 structural domains. This proteogrin seen in two form monomer and dimer both are linked by a disulfide bond. Osteoprotegerin (OPG) is encoded by TNFRSF11B genes on the cytokine receptor of tumor necrosis factor (TNF). The main function of OPG is to regulate inflammation and differentiation with R ANKL which is a transcription factor for immune-related genes [1-35].
COLONY-STIMULATING FACTORS
These are the most potent bone marrow stimulating glycoprotein binds to blood stem cells. It binds two type receptors namely granulocytes (G-CSF) and macrophages (M-CSF). These receptors are seen most commonly in the compressed zone. Both receptors help in bone remodeling process [1-37].
RECENT UPDATES ON GENETIC MANIPULATION IN TOOT MOVEMENT
Nemours experiments have been done to modulate or speed up orthodontic tooth movements. However, few recent studies reported that the administration of proteins that can stimulate osteoclasts to modulate tooth movement. The dosage related potential side effects such as root resorption are factors of consideration.
Two ways of attempt have been done, first to Administration local OPG messenger into periodontium, as shown that the inhibition of osteoclastic activity and reduce tooth movements. The gene transfer approach using a hem agglutinating virus of Japan (HVJ) envelope vector carrying mouse OPG messenger RNA (mRNA) was performed in rats for 21 days of tooth movement. The solution was injected in the mouse palatal aspect in the periodontal area. However, the study reported that no bone morphology was altered, and 50% of tooth movement was reduced. Secondly, a similar method has been RNKL messenger RNA (mRNA) using the hem agglutinating virus of Japan (HVJ) envelope vector carrying mouse. Found that 150% of osteoclastic activity has been increased.
DISCUSSION
Bone is a plastic, dynamic, and mineralized tissue that can be molded in the desired shape with a biomechanical stimulus (orthodontic forces). These unique characteristics of a bone cell initiate and propagate signaling cascade molecules critically involved in bone remodeling [1-3].
Eight of the eight articles in our study have explained that biomechanical (orthodontic forces) stimulus initiates the cascade of mechano sensing, transduction, and cellular response. This series of cascades induces the secretion of vascular endothelial growth factor (VEGF) [3-5]. VEGF factor plays a pivotal role in reducing the strain in PDL by initiating the formation of angiogenesis in bone and extracellular matrix remodeling in the periodontal ligament (PDL). VEGF factor is essential to assist local periodontal cells to differentiate, proliferate in the remodeling of gingival and alveolar bone [6-9].
Three of eight studies have reported that the receptor site to initiate the cascade of bone and soft tissue (PL, Gingiva) is present in DNA of periodontal ligament (PDL) and alveolar bone cells [1-3,6-9]. Five of the eight articles have reported that both bone and soft tissue remodeling is controlled by feedback regulatory pathway and kinas- pathway on tension and compression zone created by biomechanical stimulus [10-15].
Eight of eight studies have reported that the feedback regulatory pathway and kinas-pathway maintains the biological balance by initiating and inhibiting the process of remodeling and restoring the soft tissue. The initiation and inhibition of the feedback regulatory cascade depend on the extent of biomechanical stimulus and controlled secretion of the Tumour Necrosis Factor-alpha (TNF α) molecule in the presence of macrophage colony-stimulating factor (M-CSF) [16-25].
The response of the regulatory feedback pathway to biomechanical stimulus is to initiate osteoclast genesis during the downregulation of OPG and up-regulation of RNKL using prostaglandin E 2 (PGE 2) and interleukin (IL)-1 β synthesis in presence of M-CSF [25-36]. This process appears in the compression side within the first week of a biomechanical stimulus.
In our study, seven of eight articles have published that up-regulating of RANKL, MCSF, pathways release the signal through an inflammation-mediated nociceptive pain pathway. This process initiates within 24 h of biomechanical stimulus been applied and helps in the secretion of pro-inflammatory cytokines especially IL1 stimulates osteoclastic activity in the compressed side [37-44]. On the other hand, in the tension zone the down regulation of RNKL, an OPG pathway secretes more fibroblasts in the presence of TNF- α and reduces strain in the tension zone.
Few studies have reported that in the absence of IL-1 α and/or TNF-α signaling factor tooth movement impairs. A continuous biomechanical stimulus initiates the osteoblastic autocrine mechanism and secretes the pro-inflammatory cytokines likes IL-1 α, IL-6, IL-11, TNF- α and receptors for IL-1, IL-6, and IL-8 [37-38]. This secretion is due to the singling of osteoblast cell hence bone synthesis occurs via bone morphogenetic proteins like (a growth factor) BMP2, BMP7. This process initiates from progenitor cells like fibroblasts, osteoblasts, osteocytes, odontoblasts, in the absences of IL-1 α and/or TNF-α signaling factor within the compression side of PDL. This study concluded that light continuous or intermittent biomechanical stimulus secretes up and down the regulatory pathway for desirable tooth movement [39-43].
Six of the eight articles have been reported that in the tension zone transforming growth factor β (TGFβ) plays a pivotal role. In response to a biomechanical stimulus within the tension zone under down regulatory pathways, RANKL and M-CSF secretes new bone formation stimulating osteocytes. There are two types of TGFβ, fibroblast growth factor (FGF) and insulin-like growth factor (IGF) [44].
CONCLUSION
This systematic review gives comprehensive knowledge to readers. This study concludes that understanding the role of genes during orthodontic tooth movement is a complex process. However, this systematic review has been simplified in to three stages [45,46].
- Secretion of nociceptic pain pathway secretes neuropeptides such as Substance P, calcitonin gene related peptide (CGRP), later release vascular endothelial growth factor.
- In compression zone, up regulatory feedback RANKL, OPG play pivotal role in presence of Tumour Necrosis Factor alpha (TNF α) molecule and presence of macrophage colony-stimulating factor (M-CSF) activates osteoclast progenitors to osteoclasts in compressed side.
- In tension zone, down regulatory feedback RANKL, OPG along with transforming growth factor β (TGFβ) in presence of M-CSF and BMP2 factors activates progenitor cell to deposit bone.
This systematic review concludes answering the question that on application of biomechanical stimulus, initiates release of IL1 than activates nociceptic pain pathway followed by up and down regulatory pathways in presence of TNF α, M-CSF and TGFβ.
ACKNOWLEDGEMENT
I would like to thank Dr. Scott Anderson Editorial staff, founder editor in chief proteomics and bioinformatics, for his encouragement to re-write. However, I was working on hidden growth in orthodontic patients.
- Wirth T, Parker N, Herttuala SY (2013) History of gene therapy. Gene 525(2): 162-169.
- Drugarin D, Drugarin M, Negru S, Cioace R (2003) RANK-RANKL/OPG molecular complex—control factors in bone remodeling. TMJ 53: 296-302.
- Wu P, Chen H, Jin R, Weng T, Ho JK, et al. (2018) Non-viral gene delivery systems for tissue repair and regeneration. J Transl Med 16(1): 29.
- Karthikeyan BV, Pradeep AR (2006) Gene therapy in periodontics: A review and future implications. JCDP 7(3): 83-91.
- Kanzaki H, Chiba M, Shimizu Y, Mitani H (2002) Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor kappa B ligand up-regulation via prostaglandin E2 synthesis. J Bone Miner Res 17: 210-220.
- Cella M, Buonsanti C, Strader C, Kondo T, Salmaggi A, et al. (2003) Impaired differentiation osteoclasts in TREM-2 deficient individuals. J Exp Med 198: 645-651.
- Wilde J, Yokozeki M, Terai K, Kudo A, Moriyama K (2003) The divergent expression of periostin mRNA in the periodontal ligament during experimental tooth movement. Cell Tissue Res 312: 345-351.
- Golan I, Baumert U, Wagener H, Preising M, Lorenz B, et al. (2002) Evidence of intrafamilial variability of CBFA1/RUNX2 expression in cleidocranial dysplasia-A family study. J Orofac Orthop 63: 190-198.
- Pavlin D, Heinrich JG (2001) Effect of mechanical loading on periodontal cells. Crit Rev Oral Biol Med 12: 414-424.
- Campos MJDS, Raposo NRB, Ferreira AP, Vitral RWF (2011) Salivary alpha-amylase activity: A possible indicator of pain-induced stress in orthodontic patients. Pain Med 12(8): 1162-1166.
- Greives MR, Odessey EA, Waggoner DJ, Shenaq DS, Aradhya S, et al. (2013) RUNX2 quadruplication: Additional evidence toward a new form of syndromic craniosynostosis. J Craniofac Surg 24(1): 126-129.
- Alhashimi N, Frithiof L, Brudvik P, Bakhiet M (2000) Orthodontic movement induce high numbers of cells expressing interferon γ at mRNA and protein levels. J Interferon Cytokine Res 20: 7-12.
- Haga S, Nakaoka H, Yamaguchi T, Yamamoto K, Kim YI, et al. (2013) A genome-wide association study of third molar agenesis in Japanese and Korean populations. J Hum Genet 58: 799-803.
- Kahn AJ, Simmons DJ (1975) Investigations of cell lineage in bone using a chimera of chick and quail embryonic tissue. Nature 258: 325-327.
- Takashi N, Udagawa N, Akatsu T, Tanaka H, Shionome M, et al. (1991) Role of colony stimulating factors in osteoclast development. J Bone Min Res 6: 977-985.
- Alhashimi N, Frithiof L, Brudvik P, Bakhiet M (2004) CD 40-CD 40L expression during orthodontic tooth movement in rats. Angle Orthod 74: 100-105.
- Han D, Gong Y, Wu H, Zhang X, Yan M, et al. (2008) Novel EDA mutation resulting in X-linked non-syndromic hypodontia and the pattern of EDA-associated isolated tooth agenesis. Eur J Med Genet 51: 536-46.
- Chiba M, Takahashi I, Haruyama N, Nishimura M, Mitani H (2004) Local OPG gene transfer to periodontal tissue inhibits orthodontic tooth movement. J Dent Res 83: 920-925.
- Katagiri T, Takahashi N (2002) Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis 8: 147-159.
- Merwin JR, Anderson JM, Kocher O, Itallic CMV, Madri JA (1990) Transforming growth factor beta l modulates extracellular matrix organization and cell-cell junctional complex during in vitro angiogenesis. J Cell Physiol 142: 117-128.
- Harris EF, Butler ML (1992) Patterns of incisor root resorption before and after orthodontic correction in cases with anterior open bites. Am J Orthod Dentofacial Orthop 101: 112-119.
- Kooten CV, Banchereau J (1996) CD40-CD40 ligand a multifunctional receptor-ligand pair. Adv Immunol 61: 1-77.
- Dai J, Rabie ABM (2007) VEGF: An essential mediator of both angiogenesis and endochondral ossification. J Dent Res 86: 937-950.
- Gülden N, Eggermann T, Zerres K, Beer M, Meinelt A, et al. (2009) Interleukin-1 olymorphisms in relation to external apical root resorption (EARR). J Orofac Orthop 70: 20-38.
- Miyagawa A, Chiba M, Hayashi H, Igarashi K (2009) Compressive force induces VEGF production in periodontal tissues. J Dent Res 88: 752-756.
- Hogan BL (1996) Bone morphogenetic proteins in development. Curr Opin Genet Dev 6: 432-448.
- Quinn JM, Itoh K, Udagawa N, Hausler K, Yasuda H, et al (2001) Transforming growth factor β effects on osteoclast differentiation via direct and indirect actions. J Bone Min Res 16: 1787-1794.
- Heldin CH, Miyazono K, Dijke T (1997) TGFβ signaling from cell membrane to nucleus through SMAD proteins. Nature 319: 511-514.
- Yano S, Mentaverri R, Kanuparthi D (2005) Functional expression of α -chemokine receptors in osteoblasts: Role of regulated upon activation, normal T-cell expressed and secreted (RANTES) in osteoblasts and regulation of its secretion by osteoblasts and osteoclasts. Endocrinology 146: 2324-2335.
- Cheung WY, Liu C, Zasarsky RMLT, Simmons CA, You L (2011) Osteocyte apoptosis is mechanically regulated and induces angiogenesis in vitro. J Orthop Res 29: 523-530.
- King GN, Cochran DL (2002) Factors that modulate the effects of bone morphogenetic protein-induced periodontal regeneration: A critical review. J Periodontol 73: 925-936.
- Mitsui N, Suzuki N, Maeno M, Yanagisawa M, Koyama Y, et al. (2006) Optimal compressive force induces bone formation via increasing bone morphogenetic proteins production and decreasing their antagonist's production by Saos-2 cells. Life Sci 78(23): 2697-2706.
- Reijnders CM, Bravenboer N, Tromp AM, Blankenstein MA, Lips P (2007) Effect of mechanical loading on insulin-like growth factor-I gene expression in rat tibia. J Endocrinol 192: 131-140.
- Hirukawa K, Miyazawa K, Maeda H, Kameyama Y, Goto S, et al. (2005) Effect of tensile force on the expression of IGF-I and IGF-I receptor in organ-cultured rat cranial suture. Arch Oral Biol 50: 367-372.
- Han X, Amar S (2003) IGF-1 signaling enhances cell survival in periodontal ligament fibroblasts vs. gingival fibroblasts. J Dent Res 82: 454-459.
- Kheralla Y, Götz W, Kawarizadeh A (2010) IGF-I, IGF-IR and IRS1 expression as an early reaction of PDL cells to experimental tooth movement in the rat. Arch Oral Biol 55: 215-222.
- Bosetti M, Leigheb M, Brooks RA, Boccafoschi F, Cannas MF (2010) Regulation of osteoblast and osteoclast functions by FGF-6. J Cell Physiol 225: 466-471.
- Kawaguchi H, Chikazu D, Nakamura K, Kumegawa M, Hakeda Y (2000) Direct and indirect actions of FGF-2 on osteoclastic bone resorption in cultures. J Bone Miner Res 15: 466-473.
- Deuel TF, Kawahara RS, Mustoe TA, Pierce AF (1991) Growth factors and wound healing platelet-derived growth factor as a model cytokine. Ann Rev Med 42: 567-584.
- Sandy JR (1998) Signal transduction. Br J Orthod 25: 269-274.
- Farndale RW, Sandy JR, Atkinson SJ, Pennington SR, Meghji S, et al. (1988) Parathyroid hormone and prostaglandin E2 stimulate both inositol phosphates and cyclic Amp accumulation in mouse osteoblast cultures. Biochem J 252: 263-268.
- Davidai G, Lee A, Schvartz I, Hazum E (1992) PDGF induce tyrosine phosphorylation in osteoblast-like cells-relevance to mitogenesis. Am J Physiol 263: 205-209.
- Sandy JR, Davies M, Prime S, Farndale R (1998) Signal pathways that transduce growth factor stimulated mitogenesis in bone cells. Bone 23: 17-26.
- Safadi FF, Xu J, Smock SL, Kanaan RA, Selim AH, et al. (2003) Expression of connective tissue growth factor in bone-its role in osteoblast proliferation and differentiation in vitro and bone formation in vivo. J Cell Physiol 196: 51-62.
- Prabhakar AR, Paul JM, Basappa N (2011) Gene therapy and its implications in dentistry. Int J Clin Pediatr Dent 4(2): 85-92.
- Grol MW, Lee BH (2018) Gene therapy for repair and regeneration of bone and cartilage. Curr Opin Pharmacol 40: 59-66.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Food and Nutrition-Current Research (ISSN:2638-1095)
- Journal of Genetics and Cell Biology (ISSN:2639-3360)
- Journal of Astronomy and Space Research
- Journal of Biochemistry and Molecular Medicine (ISSN:2641-6948)
- Journal of Agriculture and Forest Meteorology Research (ISSN:2642-0449)
- Journal of Microbiology and Microbial Infections (ISSN: 2689-7660)
- Journal of Genomic Medicine and Pharmacogenomics (ISSN:2474-4670)